Stottmann Lab
The long-term goals of the Stottmann Lab are to understand the genes and molecular mechanisms guiding normal development of both the brain and face. Dr. Stottmann and his team seek to understand the genetic basis of human birth defects involving these tissues. They use a combination of human genetics and animal models to identify genes required for normal development and then study the molecular consequences of disrupting their function in the developing embryo.
Our Research Funding
- NIH/NINDS R56NS134119: Tubulin Genes in Mammalian Forebrain Development
- NIH/NINDS R01NS141189: Forward Genetic Analysis of Cortical Circuits and Structure
Join Our Team!
The Stottmann Lab is committed to mentoring the next generation of scientists and welcomes motivated undergraduate, graduate and postdoctoral researchers seeking research training opportunities. If you're interested in our work, please complete the contact form via the button below to connect with us.
Current Projects
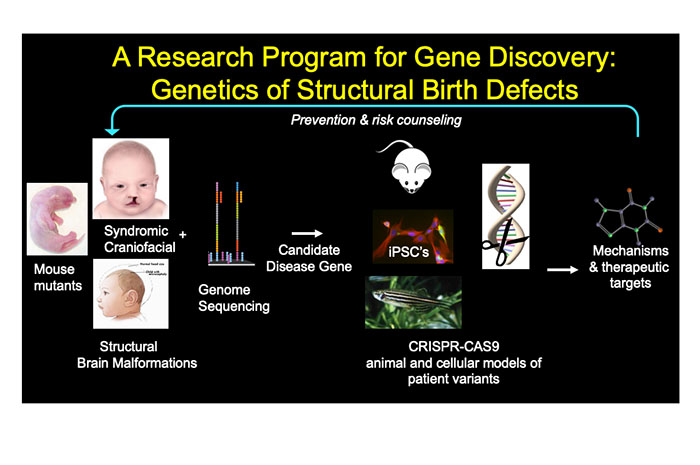
The Stottmann Lab is currently collaborating with clinical partners to directly study patients with congenital malformations of the brain and face and actively enrolling in research protocols in each of these areas. The team is using whole genome sequencing to identify potentially causal genetic variants in these patients. The lab then employs CRISPR/Cas9 genome editing technology, molecular embryology, animal models, and induced pluripotent stem cells (IPSCs) to understand how these variants lead to malformations.
As part of this work, the Stottmann Lab is pursuing a number of collaborations across the hospital and with institutions around the world. The complementary tools of genome sequencing, CRISPR genome editing, animal models and stem cell technologies offer almost limitless chances for discovery.
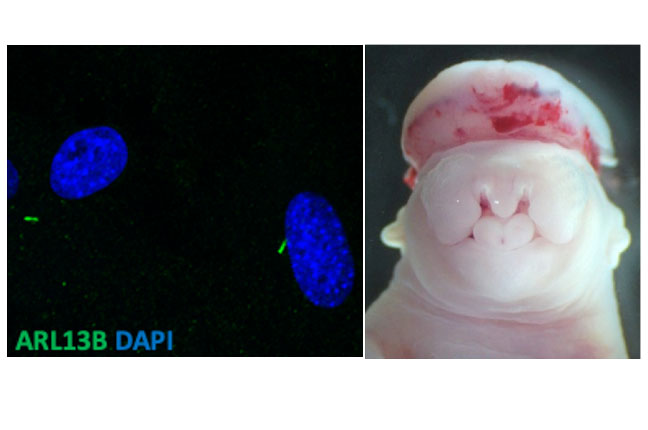
Primary cilia are organelles with a now established role in transducing many canonical developmental signaling pathways. Deficits in ciliary form and/or function lead many human conditions and are collectively called “ciliopathies.” Following the genetics behind a number of our phenotypes has led the Stottmann Lab team into the study of several genes required for normal cilia form and/or function.
The alien mutation was previously identified in a mutagenesis screen (Herron et al., Nat Genetics 2002). Positional cloning revealed this to be a null mutation in the gene Ttc21b, which is required for protein transport within the primary cilium (Tran et al., Nat Genetics 2008). The alien mutant also has a severe brain patterning defect (Stottmann et al., Dev Biol, 2009). The role of Ttc21b in the developing forebrain and craniofacial tissues, and the role of cilia in development more broadly, is a major focus within the laboratory. One direction of this project is a tissue specific ablation of Ttc21b and other primary cilia genes in the in the developing forebrain and craniofacial tissues.
Evidence from human genetics studies suggest Ttc21b is a node in a larger ciliary signaling network. The Stottmann Lab team is studying how these genes interact through studies in the mouse and in vitro. The team is also using a forward genetic approach to identify novel loci interacting with Ttc21b in mammalian development and disease and has used a Quantitative Trait Locus analysis to identify a gene it now believes at least partially explains the effects of genetic background on the alien forebrain mutation.
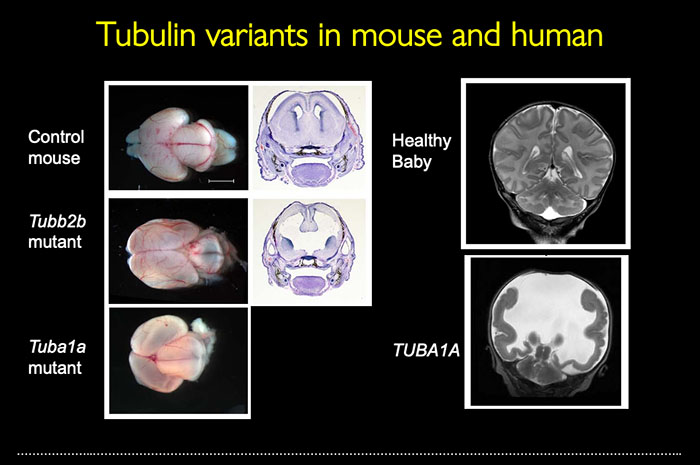
Tubulin proteins are the building blocks of microtubules, a key element of the cellular cytoskeleton. Research has shown in mice and humans that variants in tubulins lead to drastic malformations of cortical development. The tubulin genes together make up a family of genes with high-sequence similarity and, often, adjacent positions in the genome. The Stottmann Lab uses genome editing techniques to make a series of alleles to test the requirements for these genes as this has never been tested. Its team is also generating new models of human disease variants and constructing a new tool to address expression and regulation of tubulin genes.
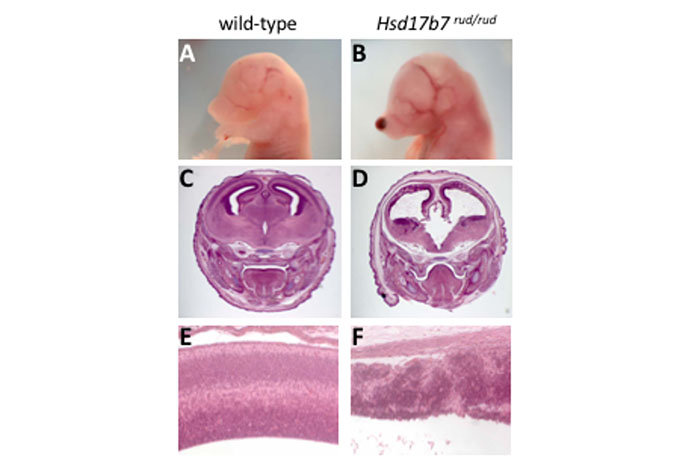
The rudolph mutation was recovered in another ENU mutagenesis screen in the Beier Lab (Stottmann et al., Genetics, 2011). Positional cloning identified an intronic mutation creating a hypomorphic allele of the gene hydroxysteroid (17-beta) dehydrogenase 7 (Hsd17b7; Stottmann et al., PLoS Genetics, 2011). Further studies of the rudolph mutation demonstrated for the first time in a genetic, in vivo, animal model a requirement for sterol metabolism in proper Sonic Hedgehog intracellular signal transduction. The rudolph mutants have a striking defect in tissue organization throughout the CNS which is not due to dysregulation of the Shh pathway. Current efforts in the lab are aimed at understanding the molecular mechanisms underlying this defect. This will contribute to a greater understanding of the role of cholesterol metabolism in cortical development.
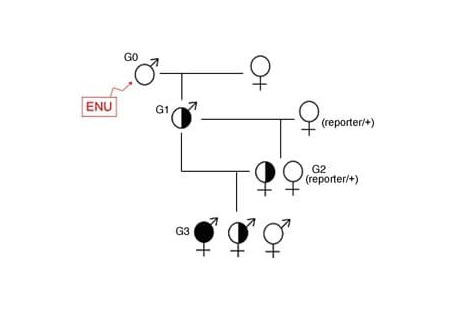
The lab primarily uses this forward genetic approach to identify genes not previously known to be required in development. They use the chemical mutagen ENU to create random mutations in the genome and breed potential mutations to homozygosity to create recessive developmental phenotypes. A combination of cutting-edge genomics and positional cloning is used to identify the mutated gene leading to the phenotypes. This turns out to be a relatively efficient tool for gene discovery in development (see Stottmann et al., 2011; Stottmann and Beier 2010). The Stottmann Lab continues to use ENU mutagenesis to create novel mutations and will increasingly employ transgenic lines to more specifically query various aspects of neuronal specification and migration. Work on this project is funded by the Cincinnati Children's Research Foundation and the National Institute of Neurological Disorders and Stroke (NINDS), part of the National Institutes of Health (NIH).
Connect With Our Team
To connect with our lab, complete the form below.



