Data Science
Our Data Science team employs innovative approaches for acquiring, managing, retrieving, and analyzing data. We employ statistical, machine learning and deep learning methods to produce predictive models for clinical decision support and population health management applications. We apply advanced information retrieval and natural language processing methods to extract knowledge from unstructured clinical notes, present that knowledge in an intuitive and user friendly fashion, and create structured predictors that can be incorporated into predictive models. Geospatial analytics allows us to employ patient and facility addresses to incorporate distance, location, socioeconomic status and consumer behavior into our predictive models. Our distributed big data infrastructure means that there are few practical limits regarding the volume, variety, and complexity of data we can analyze. Finally, taking advantage of the combined medical/dental electronic health records for our patients, we have considerable analytical experience in the Dental Informatics domain.
Deep Learning refers to a class of machine learning models that are based on large (deep) neural networks. Deep Learning facilitates the automatic learning of data representations that often lead to superior predictive models in various domains (e.g. computer vision, natural language processing, bioinformatics). We provide the capacity for state-of-the-art deep learning techniques with expertise in established neural network software libraries such as Tensorflow, Keras, or PyTorch. Our high performance computing cluster allows GPU-accelerated model optimization to tackle data at any scale.
Related Services
- Automatically learn meaningful representations directly from EHR data including medical codes, sensor data, clinical notes, insurance details, etc.
- Learning to automatically code/annotate video data, e.g. for behavioral studies
- Predictive modelling without the need of manual feature engineering
Case Study: DeepChild: Learning to predict future medical costs for individual patients
Many existing methods for patient-level risk prediction (i.e. the prediction of future medical costs) rely on expert knowledge to manually build patient representations from medical records, which is resource consuming and non-scalable. This line of work explores different deep learning models such as distributed embedding networks and attention-based recurrent networks to take up this task. These models learn patient representations based on medical claims data from over 100,000 patients across 2 years. The learned representations are predictive of patient-level medical costs in the following year and outperform various baselines. In addition to making accurate predictions, these models also allow investigation of their reasoning. For example, the attention mechanism of the recurrent network yields information about which visits and claims contribute most towards the future cost prediction.

We offer biostatistical support to pediatric dental residents and faculty in conducting dental research. We provide end-to-end statistical services from power and sample size analysis to data analysis and reporting. Our goal is to help dentists conduct high-quality cutting-edge research to achieve the highest quality of care with the lowest possible cost.
Related Services
- Statistical Consultation (e.g. sample size estimation, advice on data collection, data management and study/survey design)
- Data Manipulation (e.g., cleaning up dirty/duplicated data and data transformation)
- Data Analysis and Visualization
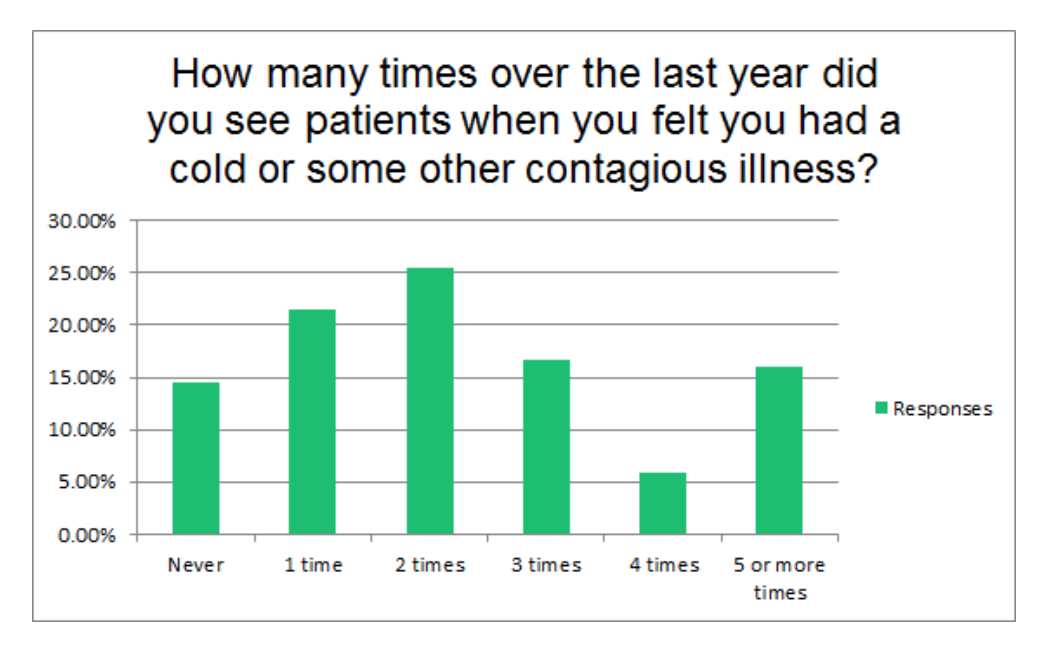
Case Study: Characteristics of Dentists Who Work While Sick
Previous studies showed that physicians working while sick result in loss of productivity and risk of spreading infections to patients. However, the phenomenon of working while sick has been minimally explored among dentists. The goal of this study is to identify risk factors and beliefs contributing to working while sick amongst practicing dentists in the United States.
We offer spatial data integration services to geocode address data to enable spatial analysis of healthcare data. Many health-related issues are related to the geographic environment we live in. By using in-house geospatial analytical software ArcGIS and Alteryx, we are able to leverage thousands of socioeconomic and demographic variables and develop state of the art geostatistical procedures to optimize health and human services. Our goal is to help you achieve better outcomes by incorporating spatial intelligence into your analytical workflow.
Related Services
- Explore disease and hazard data to test hypotheses about causes and outcomes.
- Track the spread of diseases and plan steps to combat and contain them.
- Analyze the clusters of illness among your patients to determine potential environmental causes or spread patterns.
Case Study: Evaluation of Spatial Accessibility to Ohio Trauma Centers
The goal of this study was to evaluate the spatial accessibility of Ohio trauma centers and identify potentially underserved Ohio counties. A gravity-based accessibility model using a Geographic Information System (GIS) was adapted to incorporate US census data, trauma center location data, and trauma center utilization data to quantify accessibility to trauma centers at both the zip code and county levels. An underserved index was developed to identify the location and clustering pattern of underserved regions within the state. Results suggested that most served counties were about 10 times more served than an average county while least served counties were about 4 times less served than an average county.
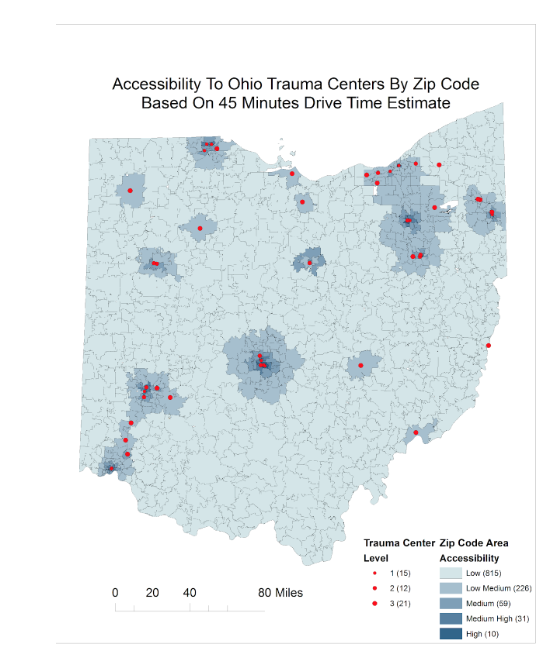
Reference: Chen, W., et al. “Evaluation of spatial accessibility to Ohio trauma centers using a GIS-based gravity model.” British Journal of Medicine and Medical Research 10.7 (2015).
The Rubix Cube is is not the only twisty puzzle, there are many more. Learn about the Pyraminx, the 2×2 and 4×4 cubes, the Megaminx on Ruwix.
Our team designs cross-platform web-based querying systems that make searching and analyzing unstructured and structured data easier, faster and more creative. We bring you the experience of instant search and data access on structured and unstructured documents with smart features such as auto suggestion, typo correction, and highlighting results. The top notch, flexible web applications designed by our Design & Technology team enable users to instantly see the results as they are typing, check distribution of data fields through charts, and save and export results in multiple formats. We also offer multi-lingual full text search with spell checking, auto-complete, highlighting, and advanced customizable ranking capabilities.
Related Services
- Search engine design
- Document ranking
Case Study #1: CardioGx
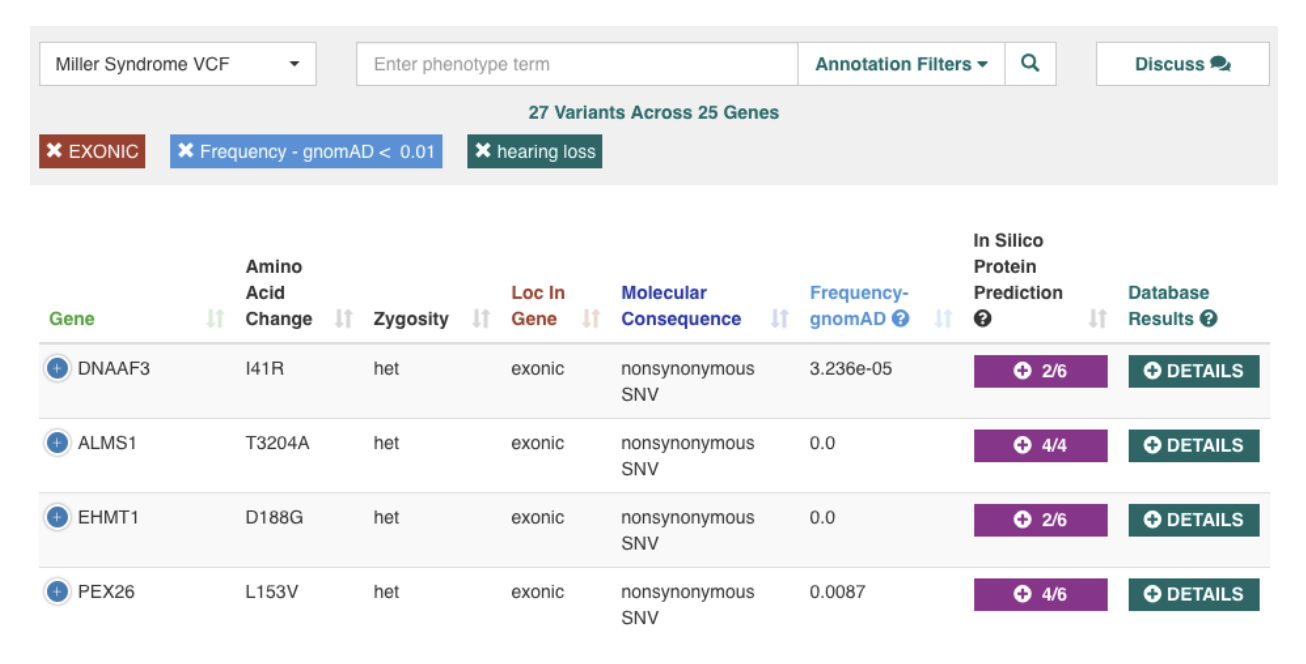
As the cost of sequencing continues to decrease, there is an upsurge in the amount of genomic data being generated, both for research as well as for clinical diagnostics. To help unlock the value in this data, we developed the CardioGenomics eXchange commons (CardioGX), a cloud-based platform with an easy-to-use interface that allows users to search genomic databases interactively, find and analyze variants, and share their analysis and findings. Users can restrict their search to look for variants with a specific frequency, pathogenicity, location in gene, and more. CardioGX enables users to upload a patient’s sequencing and phenotype data, analyze and identify causal variants, seek collaboration, and discuss genes and variants of interest with other users. System is accessible at https://cardiogx.org/
Case Study #2: QREK
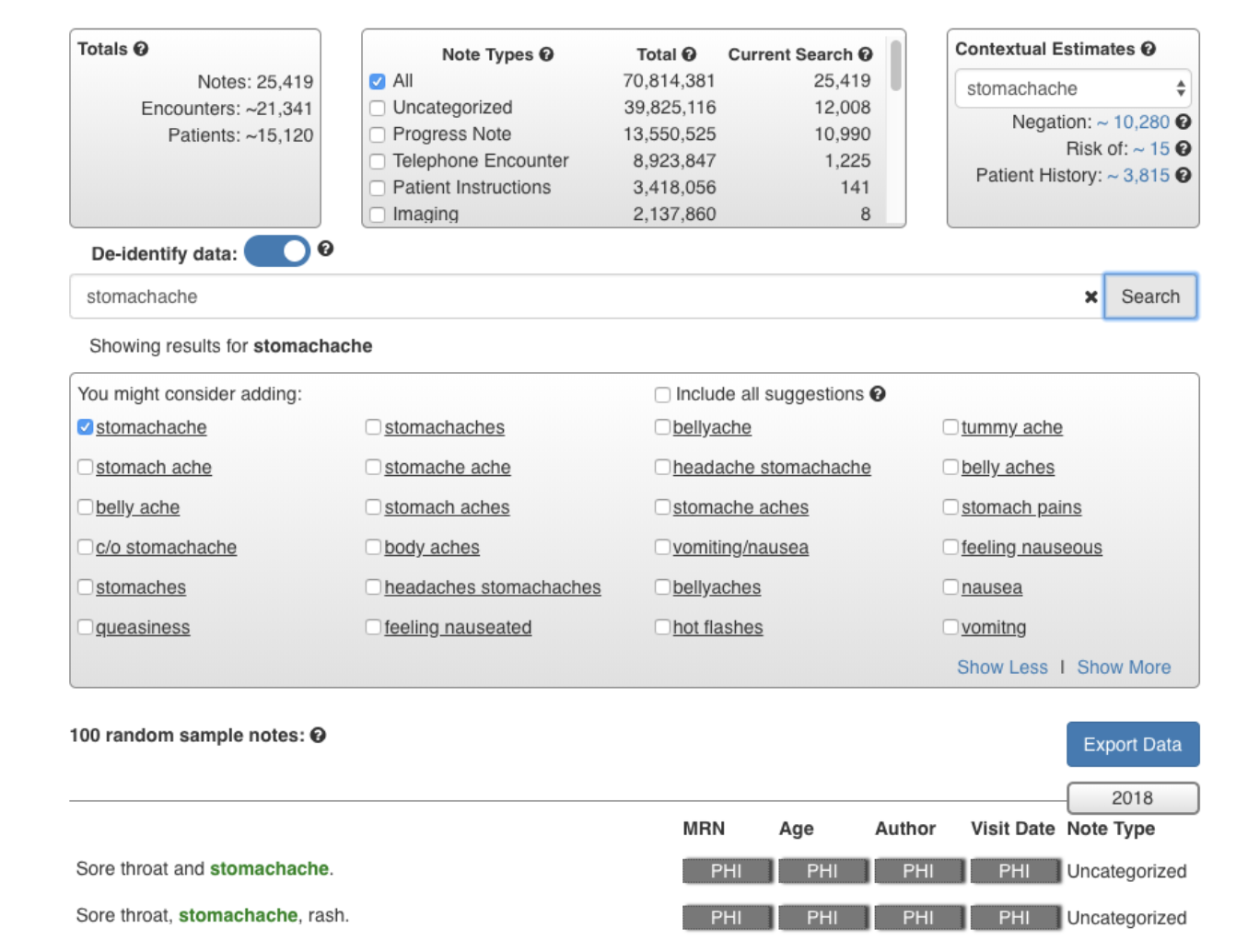
Considering more than 70% of healthcare information is stored in unstructured clinical notes, healthcare providers increasingly demand effective text-search systems for clinical care, QI reporting and research projects. However, clinical notes are known for containing spelling variations, typos, local-practice-generated acronyms, synonyms, and informal words. We developed a novel search system (QREK, also known as DeepSuggest) that can guide users to expand their input query by suggesting spelling variations, acronyms and other semantically relevant words, all identified through an artificial intelligence (AI)-driven unsupervised shallow learning algorithm, designed by our Natural Language Processing team.
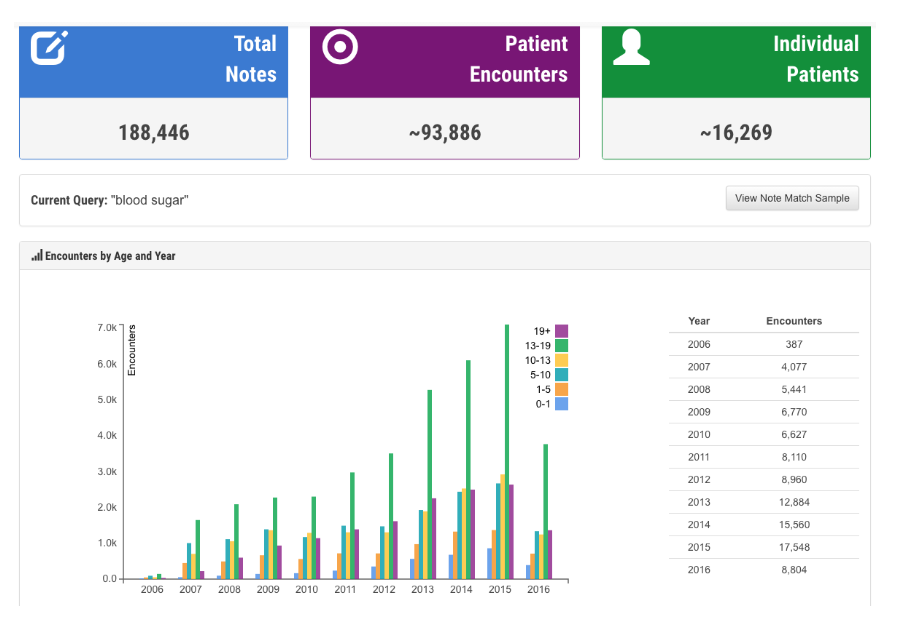
Users can filter by note type or the date notes were written. In addition to number of notes, encounters, and patients that match the search criteria, we also provide users with the distribution of note types, and an estimated number of notes with the keyword of interest being used in a context of negation, risk of, or patient history. De-identify mode allows users to use this system for demo purposes or taking screenshots, and export option enables them to export the results for further analysis. QREK was tested and validated at Nationwide Children’s Hospital with over 50 million clinical notes.
This research is funded by a Patient-Centered Outcomes Research Institute (PCORI) Award (ME-2017C1-6413). If you are interested in QREK, for access and collaboration please contact us at R&DIntake@nationwidechildrens.org
Case Study #3: Cohort Investigator
Currently most hospitals store clinical notes along with other patient information in a traditional SQL like database using various commercial systems such as EPIC. Researchers are interested to know about a cohort of patients meeting certain inclusion and/or exclusion criteria for research. Cohort Investigator is a tool that allows investigators the ability to search through both structured data and notes in order to obtain a count of patients, look through snippets of notes, view distribution charts for demographic, insurance, medication, lab and other information.
We develop Natural Language Processing (NLP) algorithms to glean data from unstructured medical records. Discrete values can be derived from millions of case narratives by an efficient and automated scan of medical notes using an NLP program. Some NLP techniques we employ include named entity recognition, topic modeling, text classification, and readability analysis, among others. With additional access to medical knowledge bases such as UMLS and SNOMED, our NLP capabilities can serve a broad range of user needs from routine text processing to evidence-based decision making.
Related Services
- Extract keyword phrases from medical narratives into discrete fields.
- Derive medical categories from text: diagnosis, symptoms, tests, and treatments.
- Identify clinical outcomes from medical notes relating to a disease such as positive vs negative cases.
Case Study: The utility of including pathology reports in improving the computational identification of Celiac patients
Celiac disease (CD) is a common autoimmune disorder. Efficient identification of patients may improve chronic management of the disease. Prior studies have shown searching International Classification of Diseases-9 (ICD-9) codes alone is inaccurate for identifying patients with CD. In this study, we developed automated classification algorithms leveraging pathology reports and other clinical data in Electronic Health Records (EHRs) to refine the subset population preselected using ICD-9 code (579.0).
EHRs were searched for established ICD-9 code (579.0) suggesting CD, based on which an initial identification of cases was obtained. In addition, laboratory results for tissue transglutaminse were extracted. Using natural language processing we analyzed pathology reports from upper endoscopy. Twelve machine learning classifiers using different combinations of variables related to ICD-9 CD status, laboratory result status, and pathology reports were experimented to find the best possible CD classifier. Ten-fold cross-validation was used to assess the results. Comparison of user-defined high-risk phrases and automated key phrases:

Reference: Chen, W., Huang, Y., Boyle, B., & Lin, S. (2016). The utility of including pathology reports in improving the computational identification of patients. Journal of pathology informatics, 7.
We employ state-of-the-art techniques drawn from artificial intelligence and statistics to build predictive models for a variety of patient outcomes, from dental caries to inpatient cardiopulmonary failure events. We work closely with our collaborators, whether healthcare providers or fellow researchers, from project inception through clinical validation to ensure the clinical decision support tools we build are precisely targeted to optimize clinical value and predictive performance.
Related Services
- Discovering patterns in large datasets.
- Data collection, visualization, and analysis.
- Statistics and machine learning expertise during study design and analysis.
Case Study #1: Predicting Pediatric Inpatient Deterioration
Unrecognized clinical deterioration outside of the pediatric intensive care unit (PICU) is an important patient safety concern. Working closely with NCH physicians and informaticians, we developed a machine learning algorithm aimed at predicting inpatient deterioration outside the PICU (i.e. code blue events, unexpected deaths, and emergency transfers to the PICU). We used lasso regularized logistic regression for model fitting and feature selection, with the final model including patient vital signs, lab results, diagnoses, and regular nursing assessment components. This predictive analytics tool–named the Deterioration Risk Index–identifies 70% more of the patients that will deteriorate within the next 24 hours than the hospital’s existing pediatric early warning system (PEWS) with no additional false alarm burden and requiring no new nursing assessments.

Case Study #2: Asthma-Related Emergency Department Recidivism
As part of Nationwide Children’s Hospital’s ‘Keep Me Well Asthma Quality Improvement Initiative,’ we teamed up with our primary care physicians and pulmonologists to predict the probability that a patient will return to the emergency department (ED) for asthma complications at the time of an asthma-related ED visit. Combining data available in a patient’s electronic health record with geospatial predictors (such as the distance between a patient’s home and our hospital), we used logistic regression to identify the top risk factors of repeated asthma-related ED visits. Using this risk-based model, our physician collaborators are now able to more effectively target patients for enrollment into existing, population-based asthma control interventions.

