Clinical Laboratory
The Steve and Cindy Rasmussen Institute for Genomic Medicine Clinical Laboratory at Nationwide Children’s Hospital performs high complexity molecular genetic analysis, cytogenetic analysis and advanced genomic testing.
The Steve and Cindy Rasmussen Institute for Genomic Medicine Clinical Laboratory also serves as the testing reference laboratory for the Acute Lymphoblastic Leukemia and Neuroblastoma Children's Oncology Groups, performing molecular and cytogenetic analyses for Children’s Oncology Group (COG) clinical trial patients.
Accreditation
The laboratory is accredited by the College of American Pathologists (CAP; #1637201 and #8162772) and certified by the Clinical Laboratory Improvement Amendments (CLIA; #36D0665271 and #36D2131848).
Faculty and Staff
The laboratory is led by eight clinical directors and one technical director. Its staff is composed of more than 30 technologists and technicians, 10 order entry and quality assurance staff, a lab operations manager, six lab supervisors, four clinical laboratory genetic counselors, five clinical bioinformatics personnel, six variant scientists and analysts, three clinical laboratory fellows and four clerical support staff.
The lab’s accessioning and quality assurance teams ensure that all clinical orders are recorded appropriately in its laboratory information management system (LIMS) and clinical reporting system. Accessioning personnel are the first people to greet you over the phone and are the receiving staff for incoming samples and testing requests. The accessioning team generates barcodes for the samples our lab receives and coordinates with the processing and testing lab sections to ensure testing is performed in a timely manner. The quality team verifies testing orders and documentation for accuracy as well as monitoring turnaround times, equipment documentation, training and competency, and procedure updates. They ensure we are following Clinical Laboratory Improvement Amendments (CLIA) guidelines and checklist requirements provided by the College of American Pathologists (CAP) as well as safety guidelines from regulatory bodies such as the United States Occupational Safety and Health Administration (OSHA).
Resources and Services
Our lab is fully equipped with instrumentation and computational resources to perform a variety of molecular, cytogenetic and advanced genomic analyses. It includes facilities for sample accessioning, tissue culture, cytogenetic analysis, fluorescence in situ hybridization (FISH), nucleic acid extraction, nucleic acid amplification, microarray analysis, methylation analysis, single gene sequencing, gene fusion analysis, next-generation sequencing and local freezer storage of patient samples.
Click here to view the guide to services, a list of all clinically offered lab tests.
A nucleic acid extraction is the act of releasing DNA/RNA from a cell in a sample for use in tests and analytical procedures.
Common Sample Types
- Blood
- Bone marrow
- Tissue biopsies
- Cheek swabs
- Saliva
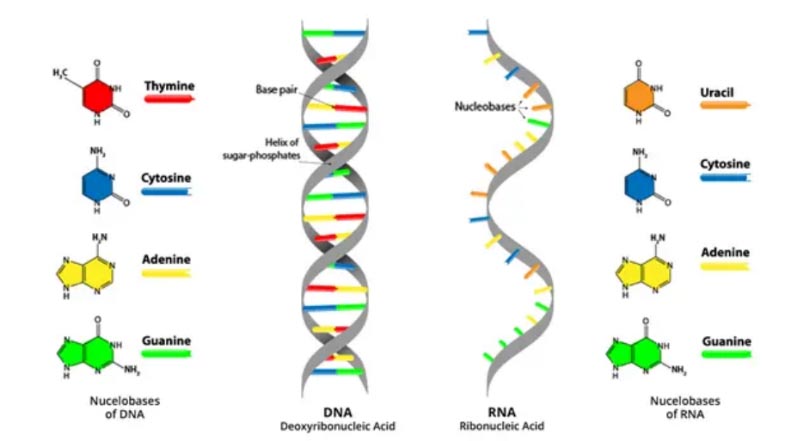
Why Is DNA/RNA Isolated?
- DNA is the blueprint, recipe or code used to make a person. These directions determine everything that make a person unique, including hair color, eye color and height.
- RNA translates the directions given by DNA so the body and its cells can read and use them.
- If there’s a typo in these directions — a missing “word” or an added “word” — the wrong information can be sent to a person’s body.
- DNA copying mistakes and “mistyped” directions can cause complications with a person's development called mutations.
- DNA and RNA are isolated during diagnostic tests and analyses to get a view of what parts of the DNA blueprint are incorrect and causing harm.
How Is DNA Isolated?
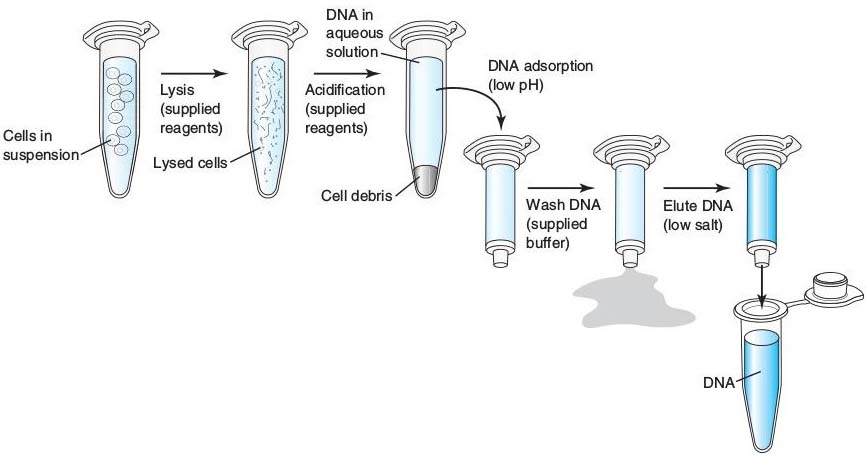
- Releasing the DNA and RNA from a cell requires breaking the cell open. The process of breaking the cell open is called cell disruption or cell lysis. This can be done through cell lysis buffer, grinding or sonicating.
- Solid‐phase extraction is commonly used for DNA isolation, concentrating the sample for analysis. In solid-phase extraction, whole blood, bone marrow or white blood cells are applied to a column in a low pH, high salt and detergent solution. The DNA, in solution, adsorbs to the solid matrix and is washed. Using a low salt solution, DNA is then released from the matrix.
Tools for Evaluating DNA/RNA Extractions
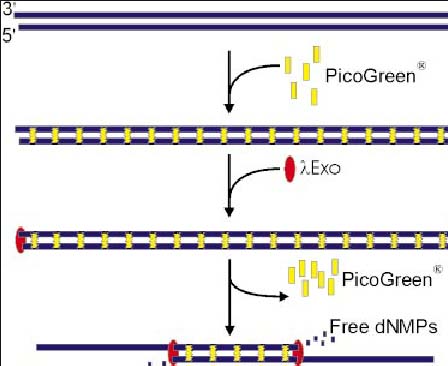
Fluorometer – Uses dyes that can be used for fluorometric quantification by binding to dsDNA or RNA specific strands.
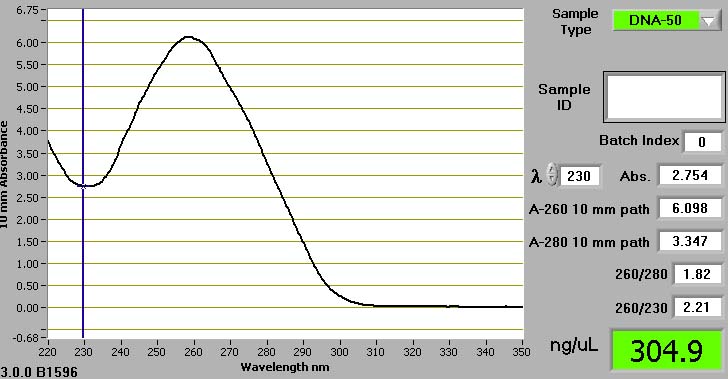
Spectrometer – Uses ultraviolet light to measure how much light is absorbed by the specimen.
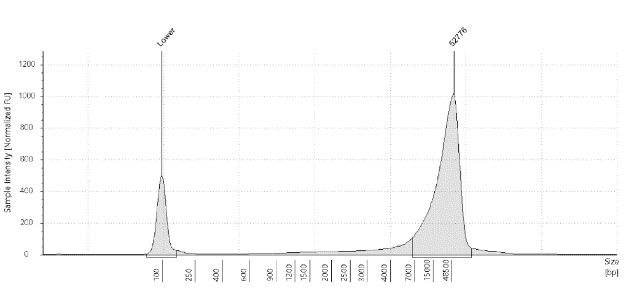
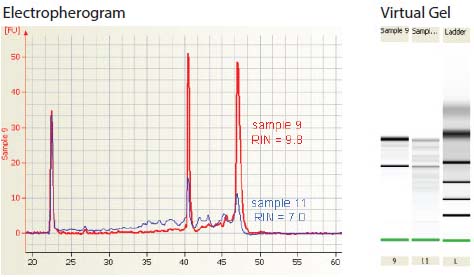
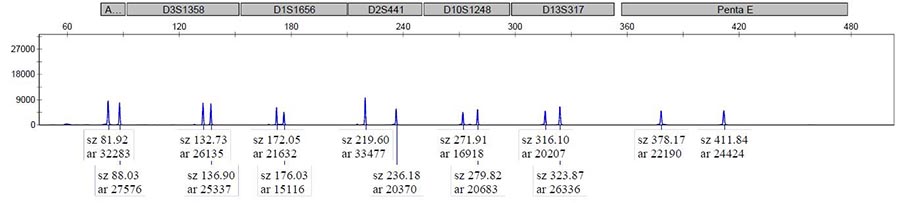
Capillary Analysis – Uses capillary electrophoresis and fluorescent detection to assigns numerical integrity values using a voltage gradient.
Gel Electrophoresis – Compares genomic weight to the ladder (a standard reference that contains DNA fragments of known lengths).
Inherited Diseases
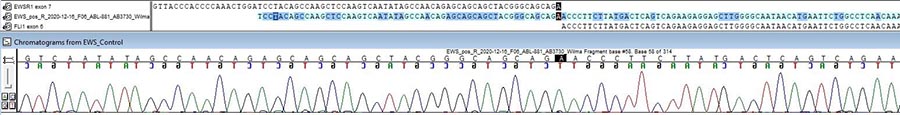
Our clinical laboratory offers numerous DNA based tests that can be used to diagnose genetic disorders and identify patients that are at risk of carrying genes associated with certain diseases. We offer a wide array of molecular tests, including sanger sequencing, fragment analysis, methylation analysis and targeted mutation analysis. Our genetic tests for inherited disease include:
- Carrier and diagnostic testing for cystic fibrosis and fragile X
- Follow up testing from newborn screening for cystic fibrosis, Krabbe disease and Hurler syndrome
- Methylation testing for Prader Willi and Angelman syndromes
- Targeted testing for hearing loss associated with Connexin26 and Connexin30 genes
- Targeted testing for known disease-associated familial variants
- Bone marrow engraftment monitoring
Cancer
Our laboratory also offers targeted oncology DNA and RNA based testing. This includes sanger sequencing in acute lymphoblastic leukemia (ALL) and central nervous system (CNS) malignancies, as well as targeted RNA fusion analysis for selected pediatric tumors. Diagnostic sequencing and screening technologies allow for more refined diagnosis and real-time identification of a subset of patients that benefit from more targeted treatment. Our genetic tests for cancer include:
- Targeted RNA sequencing analysis (RT-PCR and Archer DX)
- Targeted Sanger sequencing analysis for JAK1, JAK2, IL7R, ALK and CTNNB1
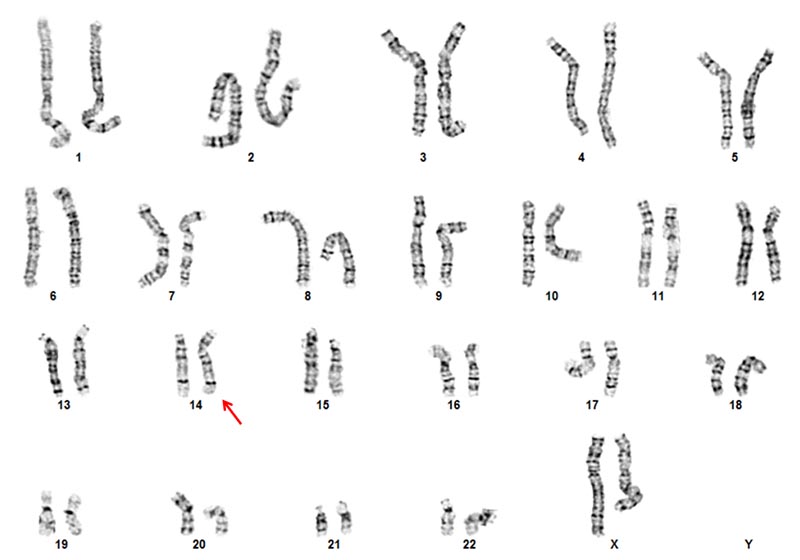
Our laboratory performs cytogenetic chromosome analysis (karyotype) to detect numerical and structural chromosomal abnormalities that may be associated with birth defects, developmental delays or disabilities, disorders of sexual differentiation, infertility, pregnancy loss and stillbirth. This test requires living cells that can grow and divide in culture. We offer chromosome studies for many sample types, including peripheral blood, skin biopsies, amniotic fluid, periumbilical blood (PUBS) samples and products of conception.
Below, karyotype image from peripheral blood studies of an individual suspected to have Down syndrome showing a Robertsonian translocation t(14;21) resulting in three copies of chromosome 21 due to chromosome rearrangement (arrow). While the majority of Down syndrome cases are due to an additional copy of chromosome 21, approximately 3% are the result of inheriting a rearrangement from a balanced carrier parent.
Fluorescence in situ Hybridization (FISH)
FISH testing is used to identify alterations within or between specific chromosome segments that are not easily visible by standard cytogenetic karyotyping. This test can be performed directly on uncultured cells, cells harvested from chromosome studies, snap frozen tissued or slides submitted from paraffin blocks. Testing involves labeling a specific chromosome target region (locus) with a fluorescent probe to look for copy number changes (losses, gains or amplifications) and other chromosome rearrangements.
Our laboratory offers a variety of FISH-based tests:
- AneuVysion for the detection of common numerical chromosome abnormalities in prenatal and products of conception (POC) samples (e.g. Trisomy 21 and Turner syndrome)
- Locus specific probes for well-known microdeletion and microduplication syndromes (e.g. DiGeorge syndrome and Prader–Willi syndrome)
- Locus specific probes for familial carrier testing
- Locus specific probes for pediatric cancers (e.g. MYCN, MYC and ALK)
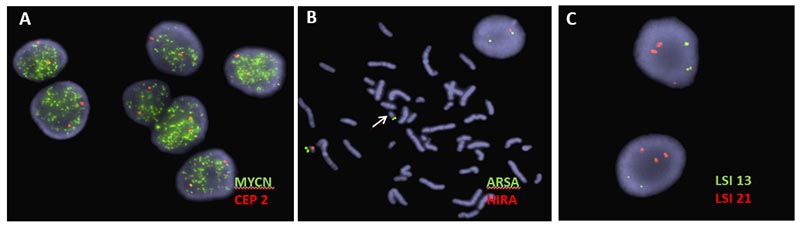
Below, examples of FISH. A) MYCN amplification shown in a snap frozen section from a neuroblastoma tumor; B) Metaphase image demonstrating loss of the DiGeorge critical region (arrow), also visible in interphase cell showing only one signal for DiGeorge probe (orange) and two signals for the control probe (green); C) Aneuvysion screen using locus specific probes for retinoblastoma (chromosome 13, green) and the Down syndrome critical region (chromosome 21, orange). Image shows fetal cells with an extra copy of chromosome 21.
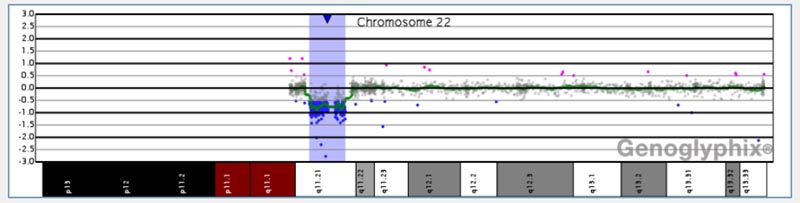
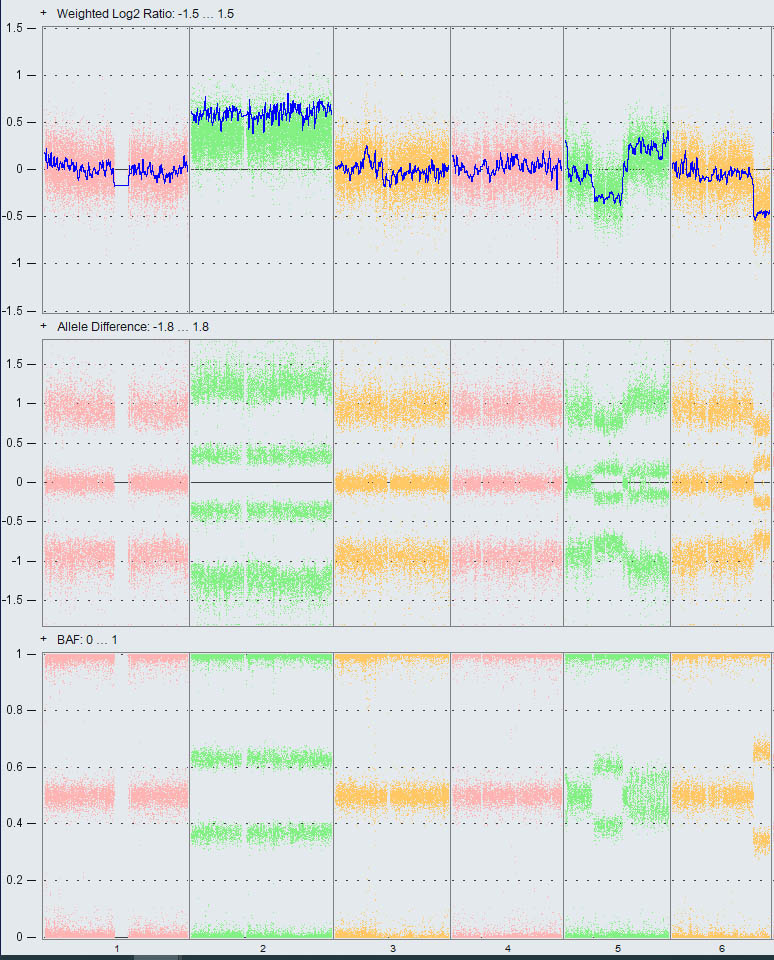
Chromosomal microarray analysis is a way to identify gains and losses of small or large parts of chromosomes by comparing the DNA from all chromosomes at once with the DNA from chromosomes of a “normal” control. This method can show chromosome gains or losses that are too small to see with standard chromosome testing through a microscope. This test also shows whether there are two unique copies of each chromosome and region.
Sometimes a person inherits two chromosomes from one parent and none from the other, resulting in large regions of homozygosity (ROH). The laboratory uses comparative genomic hybridization (CGH) for detection of copy number variants and single nucleotide polymorphism (SNP) analysis for detection of ROH. Presence of ROH is not diagnostic of any disorder, but it can suggest an increased risk for imprinting disorders and recessive genetic disorders.
Microarray analysis is frequently ordered for individuals with multiple congenital anomalies and developmental delays. It can also be performed in the prenatal setting for a fetus with ultrasound abnormalities with unknown cause.
Chromosomal microarray analysis can also be performed on cancer tissue (tumors). Our laboratory tests neuroblastoma and Wilms tumor samples for specific chromosomal gains and losses or segmental aberrations that can indicate how the tumor might behave known as prognostic indicators.
Next-generation sequencing (NGS) or massively parallel sequencing enables simultaneous analysis of multiple genes. This allows a single test to detect many different genetic disorders. Our clinical laboratory performs NGS assays to diagnose genetic disorders (Exome Sequencing, RASopathy panel) and to identify gene alterations involved in hematologic malignancies and solid tumors (Hematologic Cancer Fusion Analysis, Solid Tumor Fusion Analysis).
Exome Sequencing
Our clinical lab offers (whole) Exome-Sequencing (WES/ES) for patients with a suspected underlying genetic disorder without a clear diagnosis. ES uses next-generation sequencing to simultaneously analyze the protein coding regions of known genes in the human genome. The human genome includes protein coding DNA (exons) and non-protein coding DNA (introns). The “exome” includes exons from approximately 22,000 genes, accounting for roughly 2% of the human genome. Currently, most genetic changes (variants) known to cause genetic disorders occur within the exome.
ES sequencing can be performed on only the affected patient (proband) or the patient and biological parents. Optimal testing includes sequencing of the patient and both biological parents to determine whether variants are shared with a parent (inherited) or unique to the patient (de novo). Parental inheritance is extremely useful for identifying autosomal recessive conditions. DNA is most frequently obtained for this test from a blood sample. Less frequently, DNA may be obtained from saliva or cheek swabs.
ES is a highly complex test, requiring time for producing and analyzing NGS data. The NGS information is carefully interpreted within the context of a patient’s medical condition or phenotype by variant analysts/scientists and clinical laboratory directors.
ES can identify variants in a patient’s DNA that may be causal for a patient’s medical condition. Additionally, ES may identify variants that are unrelated to the patient’s medical condition (secondary findings) or reveal information about familial relationships. The American College of Medical Genetics and Genomics (ACMGG) has recommendations for reporting secondary findings in a specific subset of genes, which includes genes associated with inherited cancer syndromes and certain types of heart disease. Due to the complex nature of this test, genetic counseling and informed consent are essential parts of the ES testing process.
Genetic counselors are health care providers with advanced training in medical genetics and counseling. These providers help guide and support patients who wish to learn about how genetic diseases and conditions can affect them or their families.
Genetic counselors can work in different settings, including a clinical lab like this one. A lab genetic counselor works with other professionals to help ensure best patient care. They do not interact with patients.
Primary roles of genetic counselors at the Institute’s clinical lab at Nationwide Children’s include:
- Serving as a client liaison with providers by answering their questions about appropriate genetic testing
- Reaching out to providers to obtain additional information or clarify test orders
- Ensuring correct test paperwork and samples are provided to the lab
- Coordinating complex testing and ensuring all required needs are met before beginning the test
- Discussing test results with providers and how results may affect other family members of the patient
- Discussing with providers additional testing options for patients if recommended
- Educating community providers about genetic testing offered in the lab
- Helping interpret genetic test results and write lab result reports
- Arranging sending DNA and other samples to outside laboratories for genetic testing
- Working with the clinical lab team to better interpret complex test results to provide the best patient care on an individualized basis
- Helping identify appropriate labs for genetic testing done outside of the lab at Nationwide Children’s
- Helping the clinical lab bring in and implement new test offerings
- Coordinating with the research team to provide appropriate clinical testing for research patients
- Educating students, residents and lab fellows who are training in the lab
Genetic counselors in the laboratory provide support to both our clinical lab team as well as outside providers. While laboratory genetic counselors do not directly interact with patients, they assist in many ways that contribute to direct patient care.